
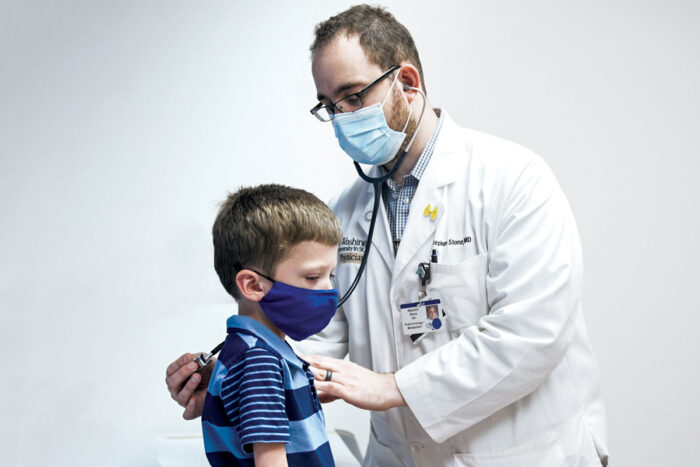
When you seek medical care, you expect a diagnosis. You may need to answer a lot of questions and undergo tests, but usually doctors can figure out the root of the problem.
This is not the case for a surprisingly large group of patients. According to the National Institutes of Health (NIH), 25 million to 30 million Americans live with rare diseases that sometimes require years to diagnose.
Funded by the NIH, the Undiagnosed Diseases Network (UDN) collaborates on hard-to-diagnose conditions in patients of all ages. With 12 clinical sites, including a coordinating site at Harvard University, the UDN joins together clinicians, researchers and physician-scientists from around the country who use advanced technologies in a way that would not be possible outside of this network.
The School of Medicine, an international leader in genome sequencing, is bringing its unique strengths to the UDN, helping to solve some of today’s most challenging medical mysteries.
In search of answers
When Amy Lair gave birth to her son, James, six weeks early, he weighed in at a healthy 8 pounds. But his growth slowed as months passed.
“He just never did really take off, as far as eating or motor skills like crawling around,” said James’ father, Jason Lair. “He was 3 years old before he walked.”
“We went through a lot of referrals from different doctors, but every test we ever did was normal,” Amy added. “Everybody said, ‘Oh, he’s fine. He’s just going to be little. He’s just a slow grower.’ As parents, we knew something was definitely wrong.”
The Lairs spent countless nights Googling symptoms and possible conditions they thought might apply to James. That’s how they came upon the UDN.
By the time families like the Lairs find out about the UDN, they have been through a “diagnostic odyssey,” said Patricia Dickson, MD, head of the Washington University UDN clinical site and the Centennial Professor of Pediatrics and professor of genetics. They have often been referred to multiple specialists who evaluate each symptom independently and try to come up with a diagnosis. But this approach often fails to explain all the patient’s underlying symptoms and issues.
“The Undiagnosed Diseases Network either gives families a way to end their diagnostic odyssey because they find an answer, or because they can be satisfied that they’ve done everything that they can and there’s nothing else left,” said Dickson, who also leads the Division of Genetics and Genomic Medicine in the Department of Pediatrics. This form of closure “is an incredibly powerful gift to families that the network provides,” she said.
Uniquely positioned
Neonatologist F. Sessions Cole, MD, led the way for Washington University’s acceptance into the UDN in 2018. Cole, the Park J. White, MD, Professor of Pediatrics, already was working on diagnosing mysterious diseases in infants. He co-leads a lab with Jennifer A. Wambach, MD, associate professor of pediatrics. This lab often is the first stop when patients from Washington University’s UDN clinical site need genetic sequencing and data analysis.
As a leader in genome sequencing since the start of the Human Genome Project, which aimed to sequence every letter of our DNA, Washington University has remained a powerhouse in the field, going on to sequence the first cancer genome and provide exceptional clinical genetics caree through its pediatric and adult genetics clinics.
Washington University also is home to one of the largest physician-scientist training programs in the country, another boon for the UDN. “We have the ability to call on clinicians from a variety of different specialties, all of whom have a research background or are actively engaged in research,” Dickson said. “They can bring both their knowledge of research and their clinical knowledge to each and every case.”
One of the UDN’s greatest assets is also one of its simplest ideas: Doctors who talk to each other, Dickson added. It’s often hard for diverse specialists to find the time and opportunity to discuss patients’ cases. But this is one of the most valuable parts of the network, said Stephen I. Stone, MD, an endocrinologist who helped with James Lair’s case.

“Collaboration is paramount in the Undiagnosed Diseases Network. Our guiding principle is solving medical mysteries through team science,” said Stone, an instructor in pediatrics, endocrinology and diabetes. After examining James, Stone was able to consult with the boy’s geneticist, Dorothy K. Grange, MD, at a UDN meeting that was set up to discuss the case. The two physicians spoke about trying growth hormone to address James’ slow growth and agreed it was the best next course of action.
A genetic coincidence
It turns out Grange, who is a professor of pediatrics in the Division of Genetics and Genomic Medicine, had another patient with the same exceptionally rare genetic change that James Lair has. This change, or variant, is in MAP2K1, a gene that makes an enzyme that is important in cell signaling. Disruption of this gene can lead to problems with cellular growth and development. The variant was not found after a genetic analysis of James’ parents, so it likely arose spontaneously in James.
When Grange first examined James, she asked Daniel J. Wegner, a lab manager in the Cole-Wambach lab, to reanalyze James’ exome sequence, which contains the sequencing data for all the protein-coding regions of the genome. After analyzing the data within their research pipeline, Wegner discovered the variant in MAP2K1, as well as two other patients with the same variant.
“By coincidence, it turned out that one of those patients was my patient that I’ve been following here at Washington University for the past 20 years,” Grange said. “I’ve been seeing her for a long, long time, trying to figure out what her diagnosis is. Without the UDN, we would not have figured this out.”
The third patient lives on the west coast and is not treated at Washington University. All three patients shared similar symptoms, with the major one being slow growth. “Finding more patients with variants in the same gene is usually the best way to solve these cases,” Wegner said.
“We have the ability to call on clinicians from a variety of different specialties.”
— Patricia Dickson, MD
Given these similarities in cases, Stone decided to prescribe growth hormone for James. Even though growth hormone has some risks, without it, “we’re looking at someone who could potentially be so short that they would have trouble living independently or without assistance,” Stone said.
One day, soon after being accepted into the UDN, Amy and Jason Lair were working on their farm with the tractor running. Amy’s phone rang. It was nurse practitioner Kathleen A. Sisco, Washington University’s UDN clinical site coordinator. She told Amy the team may have figured out what was going on with James. Amy told Jason to shut off the tractor and asked Sisco to repeat the news. “It was just a relief — instant relief,” Amy said.
“To get that news that you’ve been waiting seven years to hear — that someone has actually found something that they think is wrong with your child,” she added. “It was an amazing phone call.”
Stone added that over the past few months of being on growth hormone, James’ growth rate has been increasing quickly, and he is starting to get closer to a more normal growth curve for his age. Stone and Grange will continue to follow up with James, who is now 8 years old, and monitor his progress while he takes growth hormone.

Above and beyond
Jorge L. Granadillo, MD, an assistant professor in pediatrics for the Division of Genetics and Genomic Medicine, had a particularly tough case. Based on the symptoms he was seeing in his 7-year-old patient, it seemed very likely the boy had a genetic disorder called CHARGE syndrome. The syndrome is characterized by growth delay, hearing and vision loss and trouble communicating. But nothing in the patient’s genetic profile indicated a change in the CHD7 gene, which is known to be associated with CHARGE syndrome.
Granadillo recommended the patient apply for the UDN. Once the patient was accepted, the team got to work.
The CHD7 gene is known to play a role in epigenetics. This process regulates gene expression without modifying the gene itself. This led Granadillo’s team to do a less commonly performed epigenetic analysis called genomewide methylation analysis. The results showed methylation patterns typical of someone with CHARGE syndrome.
The team then performed whole genome sequencing, a more costly form of sequencing that is not done in a typical genetics clinic. The more detailed sequencing data further revealed a variant that was missed by traditional exome sequencing (which looks only at the protein coding regions of the genome). With some additional testing, the group demonstrated this was a new, unreported gene variant associated with CHARGE syndrome. They published their findings last year in the American Journal of Medical Genetics.
“There’s no way we could do this with all of our patients,” Granadillo said. “It requires a lot of time and resources. Resources that wouldn’t be available in a typical genetics clinic and are not covered by insurance. All of this is a clear benefit of the UDN,” he added.
Unfortunately, not all families leave with answers, said Sisco, who helps coordinate care for UDN patients. The team does make sure patients know that the UDN will continue to research their cases, particularly as new information and research findings come to light. It also provides patients with resources, such as genetic counseling, and often connects patients with other families in similar situations. “We continue to interact with these families and have ongoing relationships with them even after they leave.”
Model organisms help researchers understand rare diseases
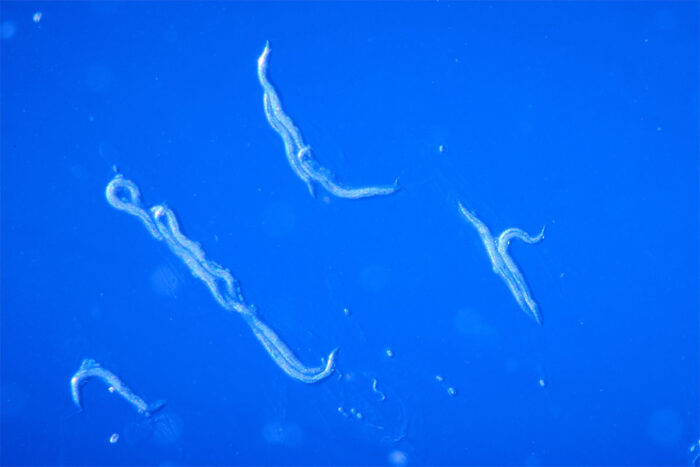
Many of the UDN cases are solved through genetic sequencing. But some cases need more investigation. The UDN has Model Organisms Screening Centers (MOSCs) that use simple organisms, such as nematode worms (C. elegans), fruit flies and zebrafish, to study complicated genetics associated with rare diseases. Washington University is home to both worm and zebrafish MOSCs.
Washington University has the only dedicated C. elegans modeling center in the country, which is co-led by Stephen C. Pak, PhD, associate professor of pediatrics, and Tim Schedl, PhD, professor of genetics. The university also is home to one of the largest zebrafish model facilities in the country, led by Lila Solnica-Krezel, PhD, who also heads the Department of Developmental Biology.
When UDN clinical team members pinpoint a genetic change or variant that potentially is causing a patient’s disease, they may send that information to the MOSCs for further study — particularly if the candidate gene is not known to cause disease.
The MOSC bioinformatics team, co-led by Dustin M. Baldridge, MD, PhD, instructor of pediatrics, determines if the candidate gene variant is appropriate for modeling by the MOSC. If so, the model organism teams decide which organism is best suited for the work.
Once the model system is determined, MOSC team members quickly learn all they can about the gene and the disease it may cause. The group often collaborates with clinicians and researchers who know about the disease or gene to better understand its mode of action.
The MOSC groups typically use advanced gene editing technology to replicate the patient’s gene variant in the model organism. The goal of the studies is to provide data in the model organism that supports the hypothesis that the candidate gene variant is indeed causing the patient’s disease. So far, the C. elegans team has provided functional support that helped diagnose 16 UDN patients, while the zebrafish team has provided support for 13 UDN diagnoses.
The MOSC team then may look for treatment strategies in that same model organism, including identifying possible drugs that might act on the gene. “These models can both provide evidence of pathogenicity and potentially be used for future studies for therapies as well,” said Angela Bowman, PhD, associate professor of developmental biology and MOSC administrative core lead.
All this work happens in a matter of months. “The team works very hard to generate the data as quickly as possible,” said Pak, the co-leader of the C. elegans core. “It’s extremely gratifying to be part of a team that helps bring diagnoses to patients and families with previously undiagnosed diseases.”

Published in the Winter 2021-22 issue


