
Together, WashU Medicine and Siteman Cancer Center have long been a powerhouse in cancer research and a national leader in providing innovative, compassionate care to patients while pushing the boundaries of what’s possible in treating this ancient and ever-evolving disease. Our researchers and clinicians are poised to shape future decades of cancer advances after having defined the previous decades of advances. And they’ll do this work in partnership with the most powerful of collaborators — their patients.
“In my experience, patients receiving cancer treatment and their family members are among the most altruistic individuals,” said Timothy J. Eberlein, MD, director of Siteman Cancer Center and the Spencer T. and Ann W. Olin Distinguished Professor and senior associate dean for cancer programs at WashU Medicine. “They often participate in research knowing it may or may not help them. But the knowledge gained from those trials will absolutely help patients in the future, which is a tremendous gift.”
That is precisely the case with two genomic tests recently developed by WashU Medicine researchers. The tests advance precision medicine approaches for treating blood cancers and solid tumors by identifying genetic changes in a patient’s cancer cells, which provides crucial information that physicians can use to help determine the optimal treatment strategy for individual patients. Both tests are approved by Medicare and Medicaid and are available to patients across the country, whether they’re treated at Siteman or another cancer center.

The tests — dubbed ChromoSeq and GatewaySeq — were developed by a team led by Eric Duncavage, MD, a professor of pathology & immunology and director of the Division of Genomic and Molecular Pathology, and David H. Spencer, MD, PhD, an associate professor of medicine and of pathology & immunology in the Division of Oncology at WashU Medicine. Both tests can identify mutations in a patient’s cancer cells and help guide treatment decisions. ChromoSeq is a whole-genome sequencing test that guides therapies for patients with blood cancers called acute myeloid leukemia (AML) and myelodysplastic syndromes. GatewaySeq identifies mutations in any solid tumor that can be targeted with FDA-approved drugs.
The development of those tests is rooted in groundbreaking research conducted at WashU Medicine just over 15 years ago involving a cancer patient at Siteman Cancer Center — a woman in her 50s — with AML that progressed rapidly.
Mapping the way to better, more personalized care
This brave patient was diagnosed in the early 2000s and spent the next few years receiving treatment at Siteman from her WashU Medicine oncologist, John F. DiPersio, MD, PhD, the Virginia E. and Sam J. Golman Endowed Professor of Medicine, director of the Center for Gene and Cellular Immunotherapy at WashU Medicine and a professor of pathology & immunology.
“She had a tremendous will to live — she fought her leukemia as hard as she could,” said her husband, who wished to maintain the family’s confidentiality. “Never said ‘Why me?’ or complained in any way. She was very interested in trying any therapy that might have a positive effect.”
During her treatment, the patient chose to participate in research studies and provided blood and skin samples for analysis. After her death from AML, when the first next-generation genome sequencing technologies became available, the research team asked whether the family would give consent for genetic sequencing of the patient’s whole cancer genome — not a collection of genes thought to be involved in cancer, but the entire DNA sequence of her cancer cells. The researchers suspected that errors in the patient’s cancer cells would provide clues to which mutations had driven her cancer to develop. Coming relatively soon after completion of the Human Genome Project, of which WashU Medicine scientists played a significant role, the sequencing of an entire cancer genome had not yet been attempted.
“We said yes,” her husband said. “Anything we could do to help the cause, we were for it.”

At the time, many were skeptical of how useful it would be to sequence a whole cancer genome. They predicted that an overwhelming number of mutations would make it impossible to single out those driving the disease. It was a risky endeavor and difficult to fund. Timothy J. Ley, MD, the Lewis T. and Rosalind B. Apple Professor of Medicine and a professor of genetics, led the project and remembers finishing about one-third of the patient’s genome and realizing they would need about $1 million more to complete the full sequence. He asked for guidance from David M. Kipnis, MD, a former head of the Department of Medicine, who had some funding ideas and made a few phone calls. One was to Alvin J. Siteman, the businessman whose generosity gave Siteman Cancer Center its name.
“That same day, we drove to Al Siteman’s office in Clayton,” Ley said. “He heard our pitch, asked probing questions, and said he’d think about it. By the time we got back to campus, his staff said we would have the funding the next day. That was the green light to go ahead. We pulled out all the stops and finished the sequencing very quickly. The analysis was difficult — we were in undiscovered territory — but we got there.”
On Nov. 6, 2008, the WashU Medicine research team published the first whole genome sequence of a cancer patient’s tumor cells in the journal Nature. The landmark study was the first to show that whole genome sequencing of a patient’s cancer could reveal gene mutations driving the disease, presenting opportunities to develop genetic tests to refine cancer diagnoses and, potentially, new therapies.
‘A watershed moment’
In November 2023, a 15-year retrospective article, also published in Nature, called that first whole cancer genome study “a watershed moment in ushering in the modern era of cancer biology and therapy.” The sequencing and analysis took over a year to complete and cost $1.6 million.
“My wife was a ‘big impact’ person in our everyday lives, so it seems fitting to me that her genome played this kind of role in cancer research,” the patient’s husband said. “We’re grateful to the researchers and their big ideas.”
The 2008 study triggered an explosion in the field of cancer genomics, leading to a revolution in the understanding of the biology of cancer. Among many critical contributions to the field, WashU Medicine investigators have led landmark studies in the genomics of breast, lung, brain, pancreatic and endometrial cancers, and many others.
Among these efforts is the development of the many genetic tests that guide doctors as they form a patient’s treatment plan, including ChromoSeq, GatewaySeq and others.
“Sequencing the first cancer genome was a phenomenal milestone in the field,” Duncavage said. “Now that genome sequencing is fast and affordable, the goal is accessibility. We want to make these tests as widely available as possible so that every patient gets molecular testing to guide their treatment from the beginning.”
Democratizing revolutionary care

Today, with ever-improving technology — much of it developed here — WashU Medicine oncologists can order the ChromoSeq test to sequence the full cancer genomes of every AML patient at Siteman who agrees to it — in a few days for a few thousand dollars — far faster, and less than 1% of the cost of the original cancer genome sequence.
Beyond leukemia, nearly all patients at Siteman have some form of genetic sequencing of their cancers. GatewaySeq — specifically designed to identify mutations in solid tumors — was developed to help oncologists match genetic mutations in a particular patient’s tumor to an ever-expanding list of FDA-approved drugs targeting those mutations.
In some cases, even if a treatable mutation is not found, sequencing can reveal how aggressive a tumor is likely to be, allowing doctors to make better decisions about the use of standard therapies — including surgery, chemotherapy, radiation therapy and, increasingly, immunotherapy.
“These tests are so important because they tell us right away if there is a mutation doctors can act on,” Duncavage said. “It has fundamentally changed the way we approach cancer therapy. We look forward to continuing this legacy, developing new methods to mine the cancer genome for future cancer therapies.”
Revolution in cancer genome sequencing defines clinical care
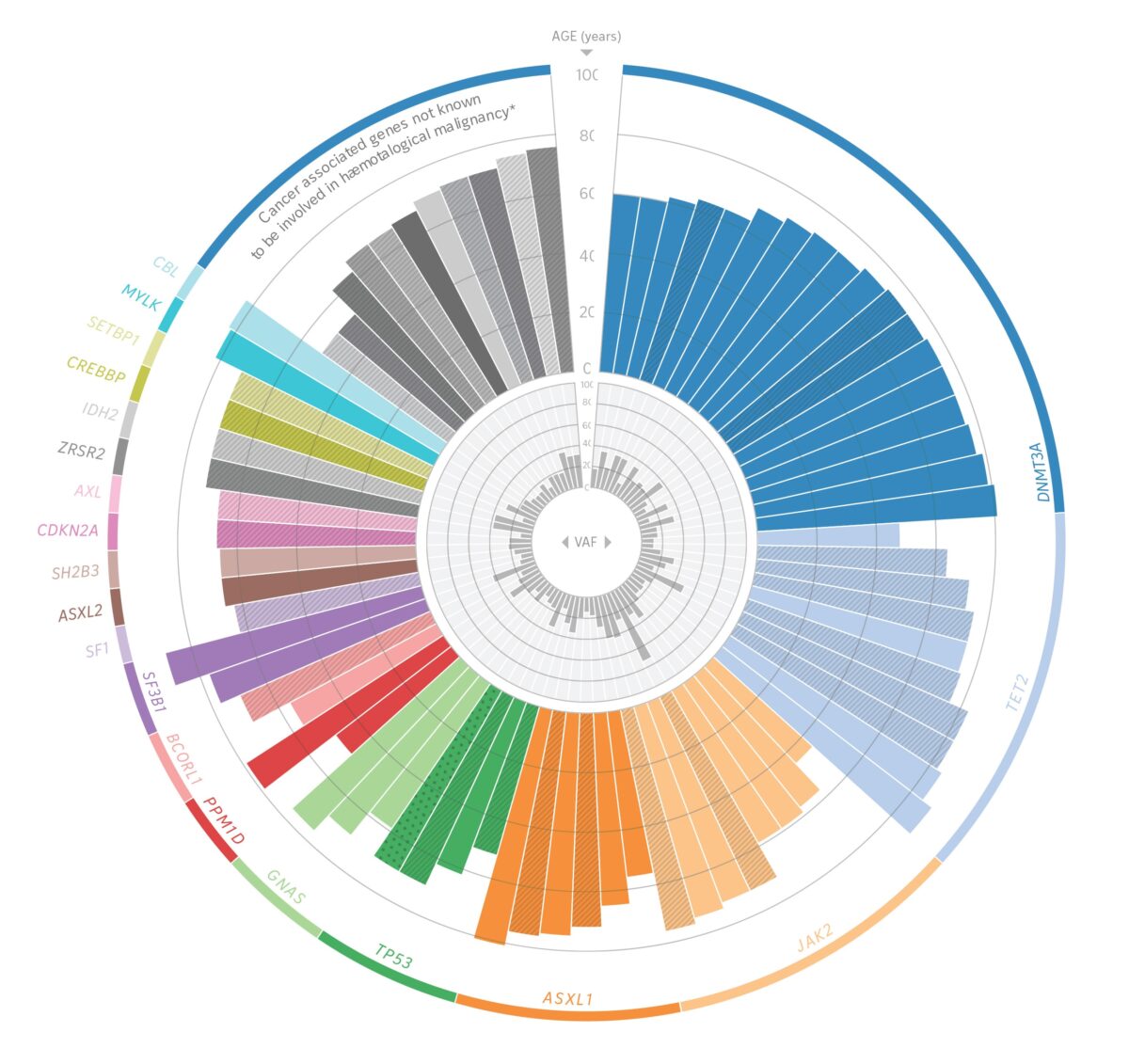
WashU Medicine was the first in the world to sequence the complete genome of a cancer patient, an endeavor that enabled the team to identify a suite of mutations driving the patient’s cancer. They also have held leadership roles in major national initiatives such as the National Cancer Institute’s Cancer Genome Atlas, in which thousands of patients with varying types of tumors have had their whole cancer genomes sequenced across dozens of tumor types. As part of these efforts, the investigators have led seminal studies in the genomics of breast, lung, brain, pancreatic and endometrial cancers, among many others.

WashU Medicine’s Li Ding, PhD, a co-author on the landmark 2008 paper that detailed the sequencing of the first cancer genome, is one of the country’s leading cancer genomics researchers. “That study was so important,” said Ding, the David English Smith Professor of Medicine and a professor of genetics. “Whole genome sequencing of tumors went from being a big unknown to a standard approach for the field of cancer research. Today, it is an increasingly important part of clinical care and helps influence decisions about the best treatment approach.”
In the ensuing years, much of the national genomics work — which Ding and her team have helped lead — has expanded beyond the genetics of tumors to include analyses of all manner of molecular changes that tumors and their surrounding environments undergo.
Most recently, Ding’s team and others have been able to use the most advanced techniques in single-cell sequencing, and so-called “omics” analyses — including proteomics, metabolomics and epigenomics — to study the 3D structures of tumors and how they may change before, during and after treatment, plus what they look like when they spread to other organs.
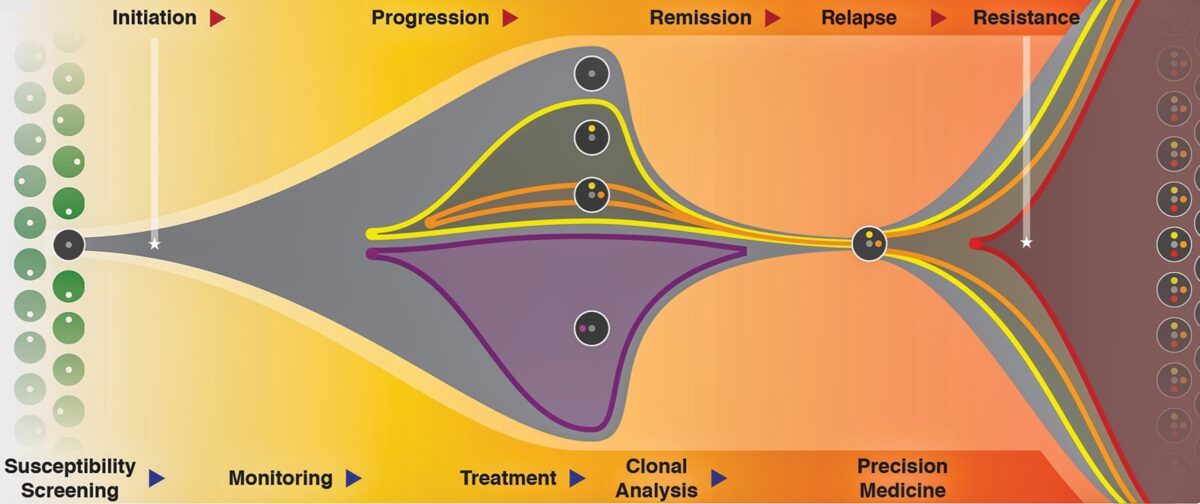
Such work continues to shape precision medicine strategies for treating cancer. As more mutations are added to genomic databases, more drugs that target specific mutations emerge. Several WashU Medicine scientists at Siteman have collaborated with pharmaceutical companies to develop these investigational therapeutics in clinical trials.
Targeted therapies for specific tumors were difficult before genetic testing. Maggie M. Mullen, MD, assistant professor of gynecologic oncology and director of the Gynecologic Oncology Tissue Bank, said today’s genetic testing panels identify drug-targetable mutations about half the time in patients with gynecologic cancers. And that proportion is likely to keep increasing.
“More and more, we are able to offer our patients targeted therapies for their specific tumors,” she said. “It’s now based much more on the molecular profile of the tumor. For example, we have a study in ovarian cancer in which each patient is assigned to one of eight drug-treatment groups based on each tumor’s specific genomic and molecular profile.”
Ding added: “It is extraordinary how far we’ve come. We hope our ongoing studies can continue revealing new insights into tumors and lead us to improve therapies and options for patients even more.”
Published in the Autumn 2024 issue


