
In early January 2020, Sean Whelan, PhD, the new head of molecular microbiology, met with Michael S. Diamond, MD, PhD, the Herbert S. Gasser Professor of Medicine. The two virologists, both active in the world of emerging infectious diseases, discussed reports of a mysterious new coronavirus that had sickened a few dozen people in the central Chinese city of Wuhan, and considered whether it was significant enough to study. No deaths had been reported.
Over the coming months, the new coronavirus would spread around the world, infecting tens of millions of people, killing more than 2 million and transforming the day-to-day lives of just about everyone. At the School of Medicine, scientists and physicians would launch themselves into the frantic worldwide effort to end the pandemic.
Research labs retooled to investigate how the virus causes disease and to develop drugs, vaccines and diagnostics to curb its destructive effects.

Informatics scientists developed models to find hot spots of infection, predict the spread of the contagion and guide public health interventions. Physicians hastily set up clinical trials to evaluate whether therapies and preventives designed for other conditions could be repurposed for COVID-19.
But all that lay in the future. In the first week of 2020, neither the virus nor the disease it caused had even been named. Whelan and Diamond still thought it possible that the outbreak would be contained, as nearly all outbreaks are, and relegated to a footnote in the history of public health.
Building the research infrastructure
By February, the novel coronavirus had spread to every province in China and a dozen countries, including the U.S. The World Health Organization had declared it a Public Health Emergency of International Concern. What was once a curiosity was shaping up to be a global pandemic on a scale not seen in over a century.
Curbing the pandemic would require information — and quickly: How is the virus getting into the body and around its defenses? Who is getting sick, and why are some people recovering while others are on ventilators? Where are the virus’s weak spots, and how could they be targeted?
Whelan teamed up with William G. Powderly, MD, director of the university’s Institute for Clinical and Translational Sciences (ICTS), and Jeffrey Milbrandt, PhD, the James S. McDonnell Professor of Genetics and executive director of the McDonnell Genome Institute, to create a COVID-19 research task force.
Whelan, also the Marvin A. Brennecke Distinguished Professor of Molecular Microbiology, took the lead on basic virology research. He coordinated Monday morning research meetings and set about establishing a high-level containment laboratory to safely study the novel coronavirus, officially named SARS-CoV-2 in February.
Because the virus is spread through the air and potentially deadly, working with it requires biosafety level-3 containment. Scientists handling the virus must wear biohazard suits with pressurized respirators, and work inside labs with multiple containment levels and specialized ventilation systems.
As an incoming department head, Whelan had been assigned ample lab space, including a room suitable for use as a biosafety level-3 laboratory. But in February, the room sat empty as Whelan’s equipment and supplies were still being shipped from his old lab in Boston. Luckily, a brand-new -80° freezer had been left in a nearby hallway by Christina L. Stallings, PhD, an associate professor of molecular microbiology, while she reorganized her space. Stallings volunteered the freezer — Whelan replaced it later — and, with Diamond’s help, outfitted the room with necessary equipment so it could be certified as a biosafety level-3 lab.
While Whelan prepared a place to work, Diamond started emailing contacts at the Centers for Disease Control and Prevention (CDC) to request access to the new coronavirus. Diamond was well known at the CDC, having conducted seminal early research during the Zika epidemic, such as creating the first pregnant mouse model and identifying protective antibodies. On Feb. 5, the CDC overnighted him a small frozen vial containing virus obtained from the first person diagnosed with COVID-19 in the U.S.
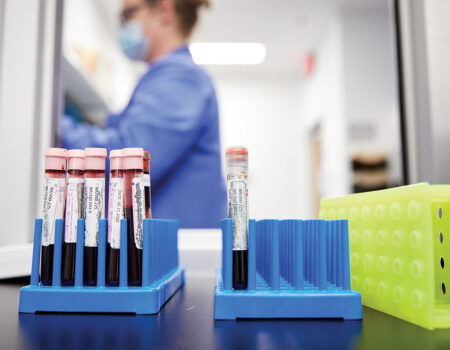
Powderly, also the J. William Campbell Professor of Medicine and the Larry J. Shapiro Director of the Institute for Public Health, coordinated COVID-19 clinical research through the ICTS. Jane A. O’Halloran, MD, PhD, an assistant professor of infectious diseases, and Philip Mudd, MD, PhD, an assistant professor of emergency medicine, already had begun creating a central biorepository of COVID-19 specimens through the School of Medicine’s Emergency Care Research Core and the Infectious Diseases Clinical Research Unit. They were collecting blood, urine and stool samples as well as nasal swabs from people diagnosed with COVID-19.

are participating in a study to evaluate large-scale COVID-19 saliva testing.
“There was a lot of concern that so many researchers would be looking for samples that it would get overwhelming for participants,” O’Halloran said. “We decided we needed to streamline the process so each patient would only be approached once and the whole community would have access to specimens.”
Powderly tapped Christina A. Gurnett, MD, PhD, a professor of neurology and the associate director of ICTS, to oversee access to the biobank. With colleagues, Gurnett reviewed nearly 200 applications from faculty in 18 medical school departments, as well as the McKelvey School of Engineering, and Arts and & Sciences. Researchers proposing similar projects were put in contact with each other to combine efforts.
“The goal wasn’t to restrict access; any request we got we tried to honor,” Gurnett said. “But these samples are a precious commodity, and we wanted to make sure we got the samples to the people who could make the best use of them right away.”
By the end of October, 15,336 samples from COVID-19 patients had been distributed to more than 23 investigative teams. O’Halloran and Mudd signed up their 500th patient Sept. 29, completing enrollment in one of the most comprehensive COVID-19 specimen collection programs in the country. They continue to collect samples for studies of the long-term effects of SARS-CoV-2 infection.
Stemming the tide
As COVID-19 continued to spread throughout the U.S. that spring, it became clear that national- and state-level models couldn’t provide the detailed information about local transmission that officials needed to make public health decisions.
“A lot of local public health agencies started developing models on their own to get locally relevant projections, and that is a heavy lift,” said Elvin H. Geng, MD, a professor of medicine who studies the effect of public health interventions on HIV infection rates. “We thought it would be helpful to develop a modeling platform that is sophisticated enough to give realistic — and publicly available — results, but simple enough that it can be accessed by an informed public health user.”
Geng and colleagues at the University of California, Berkeley, created a platform and made it publicly available on GitHub. The state of Missouri is using the platform to improve regional awareness, forecast epidemics in localities across the state and inform planning efforts. A group of hospitals in St. Louis is using the platform to project the need for hospital beds in the region.
The search for treatments
As public health officials worked to stem the tide of cases, Rachel M. Presti, MD, PhD, an associate professor of medicine and director of the university’s Infectious Diseases Clinical Research Unit, spent the spring trying to get COVID-19 patients access to experimental therapies. But deciding which trials to sign on to was no easy task.
“We reviewed lots of protocols from companies and from faculty,” Presti said. “Everyone had an idea of how to intervene. There were endless discussions about the best ways to treat people.”
The clinical research unit launched a homegrown clinical trial in the spring to test chloroquine and hydroxychloroquine as COVID-19 treatments, and was involved in some of the earliest work with blood plasma from COVID-19 survivors. They also signed on to national and international drug and vaccine clinical trials, including trials of the Janssen vaccine, a monoclonal antibody as a preventive and a different monoclonal antibody as a therapy for COVID-19.
Then, an invitation arrived to join Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV), a public-private partnership coordinated by the National Institutes of Health (NIH) to speed development of the most promising treatments and vaccines. These trials were designed to test multiple drugs at once against a single comparison group, minimizing the number of people who would receive a placebo. In addition, the trials could adapt to new data and easily add promising new drugs to the protocol.
Powderly is the head of one of the international ACTIV trials for hospitalized patients. That trial is investigating the potential of three anti-inflammatory drugs to normalize the immune response, shorten hospital stays and reduce the need for patients to be placed on ventilators to help with breathing. The goal is to enroll 2,200 patients with moderate to severe COVID-19 across the U.S. and Latin America.
In another prevention effort, a team led by Eric J. Lenze, MD, the Wallace and Lucille Renard Professor of Psychiatry, and Angela M. Reiersen, MD, associate professor of psychiatry, investigated the drug fluvoxamine for people with mild to moderate COVID-19. Fluvoxamine is used to treat obsessive-compulsive disorder, but it also regulates inflammation, which led the investigators to believe the drug could help in COVID-19. In November, Lenze and colleagues published the results of the trial in JAMA, showing that people who received the drug were less likely to require supplemental oxygen or hospitalization. A larger confirmatory trial is starting and will recruit nationwide.
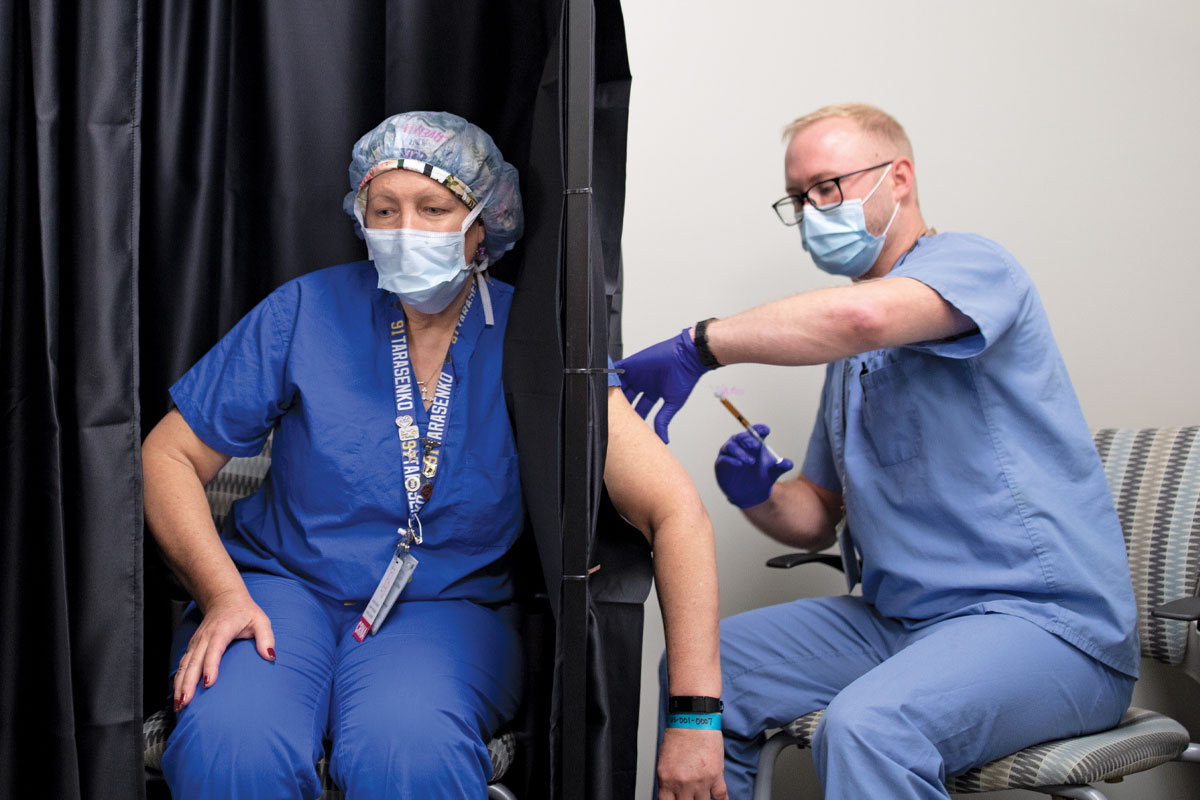
Meanwhile, Michael S. Avidan, MBBCh, the Dr. Seymour and Rose T. Brown Professor and head of the Department of Anesthesiology, was searching for ways to protect the people on the front lines from infection or at least from severe disease. With colleagues at University College London and the University of the Witwatersrand in Johannesburg, Avidan established the CROWN Coronavirus Prevention (CORONATION) trial to test the measles, mumps and rubella (MMR) vaccine. Normally given in childhood, the MMR vaccine seems to induce a generalized immune boost that protects the recipient against a wide variety of infectious diseases for several months. The trial is enrolling up to 30,000 health-care workers in Africa, Europe and North America.
Unraveling a virus
While physicians sifted through known drugs and vaccines in search of something that could prevent or treat the growing number of cases, scientists started dissecting the virus’s deadly nature to find new avenues for preventives or therapies.
First, they needed an animal model so they could study the immune response, map the disease course and test out potential drugs and vaccines in a living body. But there was a problem: Mice, the workhorses of biomedical research, are naturally resistant to SARS-CoV-2. In 2007, Carmen M. Halabi, MD, PhD, then a medical scientist training program student at the University of Iowa and now an assistant professor of pediatrics at Washington University, created a strain of genetically modified mice susceptible to infection with the SARS virus, a close cousin of SARS-CoV-2. When the COVID-19 pandemic arose, such mice became highly sought after because they were expected to be susceptible to the new coronavirus as well. But there were not nearly enough to go around.
So Diamond opted to create his own mouse model. He used a mild respiratory viral vector carrying a human gene to make mice temporarily susceptible to SARS-CoV-2 infection. Then, he infected mice with SARS-CoV-2 virus grown from the original sample shipped by the CDC. A description of this model was published in June, providing a path to studying COVID-19 for researchers who couldn’t get their hands on a genetically modified mouse.
At the same time, Diamond and Whelan started work on a vaccine using a design platform that had been successfully employed to create a vaccine for Ebola. They created a hybrid virus by swapping a gene from a mild virus with one from SARS-CoV-2, and showed that it protected mice against pneumonia by preventing infectious virus from gaining a foothold in their lungs. It is now being tested in nonhuman primates.
The hybrid virus was created as a vaccine candidate, but it could double as a research tool for scientists who lack access to a biosafety level-3 facility. It infects cells like the authentic SARS-CoV-2, but since it’s based on a harmless virus, it can be handled under ordinary laboratory safety conditions. After Diamond and Whelan published a report on their hybrid virus, requests for it poured in from around the world.
“I’ve never had this many requests for a scientific material in such a short period of time,” Whelan said. “We’ve distributed the virus to researchers in Argentina, Brazil, Mexico, Canada, the UK, Germany and, of course, all over the U.S.”
“I’ve never had this many requests for scientific material in such a short period of time.” – Sean Whelan, PhD
Diamond also worked with David T. Curiel, MD, PhD, the Distinguished Professor of Radiation Oncology, to create a vaccine that could be delivered through the nose, typically the initial site of infection. In mouse studies, Curiel and Diamond found that the nasal delivery route created a strong immune response throughout the body, but it was particularly effective in the nose and respiratory tract, preventing the infection from taking hold in the body. Later studies — in hamsters with Jacco Boon, PhD, an associate professor of medicine, and in nonhuman primates with colleagues in Montana-based NIH facilities — also were successful. If it proves as effective at preventing nasal infection in people as it is in animals, it may not only prevent infection but also curb transmission.
A few doors down, Ali H. Ellebedy, PhD, an assistant professor of pathology and immunology, was trying to generate antibodies that could be used as antiviral drugs. Using a SARS-CoV-2 protein made by Daved H. Fremont, PhD, a professor of pathology and immunology, Ellebedy injected mice, extracted antibody-producing immune cells, and then tested the antibodies’ ability to neutralize the virus.

One antibody in particular was astonishingly potent, able to cure animals at minuscule doses. But it was still a mouse antibody. If it were given to people, the human immune system would recognize it as foreign and destroy it.
And then he got an email from a name he didn’t recognize, at a biotechnology company he’d never worked with. Somehow they’d heard of his antibody, and they proposed a collaboration. Within two weeks, the company had fully “humanized” the antibody by changing its genetic sequence to trick the human immune system into thinking it was human. With the help of Boon, Ellebedy is now testing whether the humanized version retains the potency of the original mouse antibody. If it does, it could quickly enter human clinical trials as a potential antiviral.
Inadequate diagnostic testing is part of the reason the pandemic took off in the U.S.
The standard COVID-19 test involves inserting a long, thin swab uncomfortably deep into the nose. The swabs then are placed into a solution and processed. For much of this year, supplies of swabs and processing reagents were limited, making it hard to track the spread of the virus.
Milbrandt and Richard D. Head, a professor of genetics, led a highly skilled scientific team to develop a method for testing for viral RNA in saliva. The test, which received emergency use authorization from the FDA in August, requires only spitting into a cup, is highly sensitive to detecting even tiny levels of the virus, and returns a result in under a day. Washington University has adopted the saliva test to screen its undergraduates, and the state of Missouri is disseminating the testing platform statewide.
The battle continues
It has been a year since Whelan and Diamond first discussed an unnamed virus causing a tiny outbreak far away. The global pandemic still claims thousands of lives worldwide every day. And the researchers, physicians and other biomedical workers who have spent the past year battling that virus see no end in sight.
“It’s been pretty intense,” Diamond said this past fall. “I haven’t had a day off since January 2020. Not a single one. But the reward is not feeling helpless. We were the first to show intranasal vaccines work better than injectable ones. Even if our vaccine doesn’t end up being the first one, any nasal vaccine will be based on the work we did.
“A lot of people, even a lot of scientists, are feeling a pang because they can’t do anything. For us, it’s exhausting, but our labs at Washington University are making a contribution. We’re doing something. And that’s worth a lot.”
Published in the Winter 2020-21 issue


